DATA Processing
Contents
DATA Processing¶
The purpose of this Notebook is to perform the data processing steps for each sample:
DP1 : Substraction
DP2 : Gaussian fitting
One aim is to make this Notebook working within an html page and interactive using ipython widget
Workflow¶
#
To create (drag and drop image when produced)
Libraries¶
import numpy
import math
import matplotlib
import matplotlib.pyplot as plt
from matplotlib import cm
import pandas as pd
from glob import glob
from functools import reduce
import ipywidgets as widgets
import matplotlib.gridspec as gridspec
from ipywidgets import interact
import re
from cycler import cycler
import seaborn as sns
import matplotlib.gridspec as gridspec
import pylab
from lmfit.models import ExponentialModel, GaussianModel
%matplotlib
Using matplotlib backend: Qt5Agg
0 Input parameters¶
Not use (but here just in case)
Take time to describe method used with respect to previous Notebook (Improvement - Emphase on python teaching)
Sample type¶
# 0.1 Date
date = "2020_09_16"
# 0.2 XP
# Can take value XP_1-1, XP_1-2
XP = "XP_1-1"
# 0.3 Sample type
# Can take value ASW, C2H6, C2H6_ASW
spl = "ASW"
Sample annex¶
For quick sample comparison (max 3 sample named 2-3-4)
date2 = "2020_09_28"
date3 = "2020_09_17"
date4 = "2020_09_21"
\(\color{green}{\text{DP1 : Sample Checking}}\) ¶
1.1 DATA Import¶
The Data processing routine is performed per sample.
A date is set as input
from the previous data analysis, summarized in the following document below, we have identified 4 temeperature ranges where the sample undergo different physical processes.
XP1_Plan
So the purpose here is to chop the data with respect to those ranges before to perform the Substraction
Experimental ranges (in T):
Range 1: 20 - 100K
Range 2: 100 to 130K
Range 3: 130K - 140K
Range 4: Beyond 140K
I want to extract the temperature ramp info from XP Ramp:
1.1.1 XP-Ramp¶
XP_Ramp_df = pd.read_csv('D:\DATA-Processing\PAC\XP_list_test.csv')
XP_Ramp_df_I = XP_Ramp_df.set_index('Date')
#XP_Ramp_df_I.head(5)
1.1.2 DR1, DR2, DR3¶
DR1 (Date)¶
DR1_Allscan_df = pd.read_csv("D:\DATA-Processing\PAC\{}/Samples/{}/Data/DR/DR1_{}_All-scans.csv".format(XP,date,date))
DR1_Allscan_df = DR1_Allscan_df.T.iloc[1:].T
#DR1_Allscan_df
DR1 (Date Annex)¶
DR1_Allscan_df_2 = pd.read_csv("D:\DATA-Processing\PAC\{}/Samples/{}/Data/DR/DR1_{}_All-scans.csv".format(XP,date2,date2))
DR1_Allscan_df_3 = pd.read_csv("D:\DATA-Processing\PAC\{}/Samples/{}/Data/DR/DR1_{}_All-scans.csv".format(XP,date3,date3))
DR1_Allscan_df_4 = pd.read_csv("D:\DATA-Processing\PAC\{}/Samples/{}/Data/DR/DR1_{}_All-scans.csv".format(XP,date4,date4))
DR1_Allscan_df_2 = DR1_Allscan_df_2.T.iloc[1:].T
DR1_Allscan_df_3 = DR1_Allscan_df_3.T.iloc[1:].T
DR1_Allscan_df_4 = DR1_Allscan_df_4.T.iloc[1:].T
#DR1_Allscan_df_2
DR2 (Date)¶
DR2_Allscan_df = pd.read_csv('D:\DATA-Processing\PAC\{}/Samples/{}/Data/DR/DR2_{}_All-scans.csv'.format(XP, date, date))
DR2_Allscan_df = DR2_Allscan_df.T.iloc[1:].T
#DR2_Allscan_df
DR2 (Date Annex)¶
DR2_Allscan_df_2 = pd.read_csv('D:\DATA-Processing\PAC\{}/Samples/{}/Data/DR/DR2_{}_All-scans.csv'.format(XP, date2, date2))
DR2_Allscan_df_3 = pd.read_csv('D:\DATA-Processing\PAC\{}/Samples/{}/Data/DR/DR2_{}_All-scans.csv'.format(XP, date3, date3))
DR2_Allscan_df_4 = pd.read_csv('D:\DATA-Processing\PAC\{}/Samples/{}/Data/DR/DR2_{}_All-scans.csv'.format(XP, date4, date4))
DR2_Allscan_df_2 = DR2_Allscan_df_2.T.iloc[1:].T
DR2_Allscan_df_3 = DR2_Allscan_df_3.T.iloc[1:].T
DR2_Allscan_df_4 = DR2_Allscan_df_4.T.iloc[1:].T
#DR2_Allscan_df_2
DR3 (Date)¶
DR3_Allscan_df = pd.read_csv("D:\DATA-Processing\PAC\{}/Samples/{}/Data/DR/DR3_{}_A.csv".format(XP,date,date))
DR3_Allscan_df = DR3_Allscan_df.T.iloc[1:].T
#DR3_Allscan_df
DR3 (Date Annex)¶
DR3_Allscan_df_2 = pd.read_csv("D:\DATA-Processing\PAC\{}/Samples/{}/Data/DR/DR3_{}_A.csv".format(XP,date2,date2))
DR3_Allscan_df_3 = pd.read_csv("D:\DATA-Processing\PAC\{}/Samples/{}/Data/DR/DR3_{}_A.csv".format(XP,date3,date3))
DR3_Allscan_df_4 = pd.read_csv("D:\DATA-Processing\PAC\{}/Samples/{}/Data/DR/DR3_{}_A.csv".format(XP,date4,date4))
DR3_Allscan_df_2 = DR3_Allscan_df_2.T.iloc[1:].T
DR3_Allscan_df_3 = DR3_Allscan_df_3.T.iloc[1:].T
DR3_Allscan_df_4 = DR3_Allscan_df_4.T.iloc[1:].T
1.3 Data_annex¶
1.2.1 Import¶
#Date
data_anex_df = pd.read_csv("D:\DATA-Processing\PAC\{}/Samples/{}/Data/DR/{}_data_annex.csv".format(XP,date,date))
#Date Annex
data_anex_df_2 = pd.read_csv("D:\DATA-Processing\PAC\{}/Samples/{}/Data/DR/{}_data_annex.csv".format(XP,date2,date2))
data_anex_df_3 = pd.read_csv("D:\DATA-Processing\PAC\{}/Samples/{}/Data/DR/{}_data_annex.csv".format(XP,date3,date3))
data_anex_df_4 = pd.read_csv("D:\DATA-Processing\PAC\{}/Samples/{}/Data/DR/{}_data_annex.csv".format(XP,date4,date4))
data_anex_df
| Name | min1 | index1 | min2 | index2 | min3 | index3 | min4 | index4 | min5 | ... | maxA0w | maxBi | maxB | maxBw | maxCi | maxC | maxCw | Int_A | Int_C | Int_N_A | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | ASW_2020_09_16_1 | -0.018844 | 6636 | -0.008464 | 4235 | -0.008369 | 4139 | -0.001418 | 2403 | -0.000786 | ... | 3268.321 | 2945 | 0.009284 | 2219.700 | 1773 | 0.023557 | 1654.651 | 95.913079 | 6.952239 | 100.000000 |
| 1 | ASW_2020_09_16_2 | -0.017376 | 6636 | -0.006804 | 4235 | -0.006714 | 4139 | 0.000216 | 2432 | 0.000863 | ... | 3264.464 | 2945 | 0.009251 | 2219.700 | 1771 | 0.023602 | 1653.686 | 96.271574 | 7.064048 | 100.373772 |
| 2 | ASW_2020_09_16_3 | -0.015208 | 6636 | -0.005247 | 4197 | -0.005165 | 4130 | 0.001310 | 2413 | 0.001796 | ... | 3264.464 | 2947 | 0.010343 | 2220.665 | 1754 | 0.024362 | 1645.490 | 98.372535 | 7.669780 | 102.564255 |
| 3 | ASW_2020_09_16_4 | -0.014749 | 6631 | -0.004931 | 4197 | -0.004867 | 4130 | 0.001626 | 2413 | 0.002152 | ... | 3264.464 | 2947 | 0.010512 | 2220.665 | 1754 | 0.024289 | 1645.490 | 98.949916 | 7.710412 | 103.166239 |
| 4 | ASW_2020_09_16_5 | -0.014274 | 6620 | -0.004424 | 4245 | -0.004362 | 4130 | 0.002212 | 2413 | 0.002702 | ... | 3264.464 | 2947 | 0.010562 | 2220.665 | 1754 | 0.024293 | 1645.490 | 99.128959 | 7.740047 | 103.352911 |
| 5 | ASW_2020_09_16_6 | -0.013616 | 6636 | -0.003845 | 4235 | -0.003754 | 4139 | 0.003361 | 2444 | 0.003725 | ... | 3255.303 | 2949 | 0.010078 | 2221.629 | 1756 | 0.023139 | 1646.454 | 102.427355 | 8.047927 | 106.791854 |
| 6 | ASW_2020_09_16_7 | -0.013178 | 6632 | -0.003492 | 4235 | -0.003383 | 4139 | 0.003972 | 2425 | 0.004340 | ... | 3255.303 | 2951 | 0.010252 | 2222.593 | 1756 | 0.022505 | 1646.454 | 103.570509 | 8.057326 | 107.983719 |
| 7 | ASW_2020_09_16_8 | -0.012907 | 6632 | -0.003190 | 4235 | -0.003086 | 4139 | 0.004323 | 2406 | 0.004697 | ... | 3255.303 | 2951 | 0.010345 | 2222.593 | 1756 | 0.022478 | 1646.454 | 103.920588 | 8.069566 | 108.348714 |
| 8 | ASW_2020_09_16_9 | -0.013285 | 6636 | -0.002969 | 4245 | -0.002883 | 4139 | 0.004149 | 2415 | 0.004730 | ... | 3251.446 | 2959 | 0.011390 | 2226.450 | 1754 | 0.022548 | 1645.490 | 106.316388 | 8.023931 | 110.846602 |
| 9 | ASW_2020_09_16_10 | -0.013301 | 6636 | -0.002779 | 4216 | -0.002666 | 4139 | 0.004334 | 2367 | 0.004952 | ... | 3251.446 | 2959 | 0.011787 | 2226.450 | 1754 | 0.022958 | 1645.490 | 107.623907 | 8.026164 | 112.209835 |
| 10 | ASW_2020_09_16_11 | -0.013018 | 6636 | -0.002502 | 4197 | -0.002379 | 4120 | 0.004644 | 2367 | 0.005298 | ... | 3250.482 | 2959 | 0.011822 | 2226.450 | 1754 | 0.022904 | 1645.490 | 107.980853 | 8.037065 | 112.581991 |
| 11 | ASW_2020_09_16_12 | -0.012113 | 6636 | -0.002024 | 4149 | -0.001986 | 4130 | 0.005265 | 2415 | 0.005871 | ... | 3251.446 | 2959 | 0.011693 | 2226.450 | 1754 | 0.020700 | 1645.490 | 108.386906 | 8.033410 | 113.005345 |
| 12 | ASW_2020_09_16_13 | -0.011900 | 6636 | -0.001360 | 4151 | -0.001341 | 4132 | 0.005725 | 2367 | 0.006299 | ... | 3250.482 | 2980 | 0.012006 | 2236.575 | 1771 | 0.021517 | 1653.686 | 109.276611 | 8.503723 | 113.932962 |
| 13 | ASW_2020_09_16_14 | -0.011673 | 6636 | -0.001157 | 4151 | -0.001115 | 4130 | 0.005912 | 2415 | 0.006504 | ... | 3250.482 | 2959 | 0.011885 | 2226.450 | 1771 | 0.021516 | 1653.686 | 109.502264 | 8.477687 | 114.168230 |
| 14 | ASW_2020_09_16_15 | -0.011645 | 6636 | -0.001251 | 4149 | -0.001226 | 4139 | 0.006022 | 2415 | 0.006723 | ... | 3250.482 | 2966 | 0.011713 | 2229.825 | 1699 | 0.020339 | 1618.973 | 109.378492 | 8.007280 | 114.039184 |
| 15 | ASW_2020_09_16_16 | -0.011338 | 6636 | -0.000905 | 4168 | -0.000901 | 4139 | 0.006207 | 2415 | 0.006852 | ... | 3247.107 | 2964 | 0.012078 | 2228.861 | 1769 | 0.020189 | 1652.722 | 109.838625 | 8.093852 | 114.518923 |
| 16 | ASW_2020_09_16_17 | -0.011244 | 6636 | -0.000821 | 4149 | -0.000807 | 4139 | 0.006330 | 2415 | 0.006958 | ... | 3247.107 | 2964 | 0.012166 | 2228.861 | 1769 | 0.020213 | 1652.722 | 110.041893 | 8.142568 | 114.730852 |
| 17 | ASW_2020_09_16_18 | -0.011332 | 6636 | -0.000989 | 4149 | -0.001025 | 4130 | 0.006762 | 2480 | 0.007738 | ... | 3246.625 | 2957 | 0.011663 | 2225.486 | 1754 | 0.021377 | 1645.490 | 109.763420 | 7.476483 | 114.440514 |
| 18 | ASW_2020_09_16_19 | -0.010872 | 6636 | -0.000882 | 4158 | -0.000914 | 4130 | 0.006530 | 2480 | 0.008113 | ... | 3246.625 | 2978 | 0.011855 | 2235.610 | 1697 | 0.021481 | 1618.009 | 109.666245 | 7.569996 | 114.339198 |
| 19 | ASW_2020_09_16_20 | -0.010809 | 6636 | -0.000910 | 4149 | -0.000942 | 4130 | 0.006549 | 2480 | 0.008185 | ... | 3246.143 | 2978 | 0.011966 | 2235.610 | 1697 | 0.021457 | 1618.009 | 109.738473 | 7.560006 | 114.414504 |
| 20 | ASW_2020_09_16_21 | -0.011159 | 6636 | -0.001222 | 4151 | -0.001287 | 4103 | 0.006266 | 2415 | 0.007035 | ... | 3242.286 | 2966 | 0.012110 | 2229.825 | 1699 | 0.020884 | 1618.973 | 109.665610 | 8.124397 | 114.338537 |
| 21 | ASW_2020_09_16_22 | -0.010915 | 6630 | -0.001349 | 4151 | -0.001461 | 4111 | 0.006305 | 2444 | 0.007058 | ... | 3232.161 | 2976 | 0.012184 | 2234.646 | 1535 | 0.020529 | 1539.905 | 109.891203 | 8.369954 | 114.573742 |
| 22 | ASW_2020_09_16_23 | -0.010836 | 6630 | -0.001540 | 4151 | -0.001650 | 4084 | 0.006335 | 2396 | 0.007176 | ... | 3228.304 | 2976 | 0.012485 | 2234.646 | 1535 | 0.020793 | 1539.905 | 110.395286 | 8.440394 | 115.099305 |
| 23 | ASW_2020_09_16_24 | -0.014164 | 6636 | -0.006225 | 4149 | -0.006255 | 4101 | 0.003633 | 2444 | 0.004862 | ... | 3228.304 | 2976 | 0.011491 | 2234.646 | 1697 | 0.019248 | 1618.009 | 108.090993 | 7.265889 | 112.696823 |
| 24 | ASW_2020_09_16_25 | -0.013780 | 6636 | -0.008598 | 4151 | -0.008599 | 4122 | 0.001659 | 2484 | 0.002442 | ... | 3228.304 | 2978 | 0.008953 | 2235.610 | 1535 | 0.016275 | 1539.905 | 83.840839 | 5.785232 | 87.413354 |
| 25 | ASW_2020_09_16_26 | 0.002459 | 6636 | 0.004937 | 4151 | 0.004895 | 4053 | 0.006344 | 2329 | 0.006488 | ... | 3227.822 | 2978 | 0.004050 | 2235.610 | 1535 | 0.009392 | 1539.905 | 27.229288 | 2.969525 | 28.389547 |
| 26 | ASW_2020_09_16_27 | 0.007303 | 6636 | 0.007753 | 4533 | 0.008098 | 3959 | 0.007518 | 2300 | 0.007247 | ... | 3228.304 | 2295 | 0.001530 | 1906.319 | 1495 | 0.004807 | 1520.620 | 3.170572 | 1.399266 | 3.305672 |
| 27 | ASW_2020_09_16_28 | 0.007890 | 6332 | 0.008135 | 4545 | 0.008503 | 4139 | 0.008035 | 2480 | 0.007282 | ... | 3242.286 | 2698 | 0.001674 | 2100.615 | 1495 | 0.006589 | 1520.620 | 2.966754 | 1.841489 | 3.093169 |
| 28 | ASW_2020_09_16_29 | 0.008264 | 6332 | 0.008545 | 4563 | 0.008896 | 4139 | 0.008449 | 2480 | 0.007719 | ... | 3242.286 | 2698 | 0.001720 | 2100.615 | 1495 | 0.006580 | 1520.620 | 3.023007 | 1.840784 | 3.151819 |
| 29 | ASW_2020_09_16_30 | 0.008619 | 6331 | 0.008641 | 4562 | 0.009109 | 4084 | 0.008596 | 2480 | 0.008381 | ... | 3228.304 | 2602 | 0.001573 | 2054.332 | 1574 | 0.004599 | 1558.708 | 3.025809 | 1.338555 | 3.154741 |
| 30 | ASW_2020_09_16_31 | 0.008318 | 6636 | 0.008889 | 4543 | 0.009277 | 3959 | 0.008351 | 2461 | 0.008204 | ... | 3223.965 | 2437 | 0.002416 | 1974.781 | 1495 | 0.006633 | 1520.620 | 3.256800 | 1.924223 | 3.395574 |
31 rows × 31 columns
#data_anex_df = data_anex_df.T.iloc[1:].T
#data_anex_df_2 = data_anex_df_2.T.iloc[1:].T
#data_anex_df_3 = data_anex_df_3.T.iloc[1:].T
#data_anex_df_4 = data_anex_df_4.T.iloc[1:].T
data_anex_df_2
| Name | min1 | index1 | min2 | index2 | min3 | index3 | min4 | index4 | min5 | ... | maxA0w | maxBi | maxB | maxBw | maxCi | maxC | maxCw | Int_A | Int_C | Int_N_A | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | ASW_2020_09_28_1 | -0.026504 | 6620 | -0.024226 | 4401 | -0.022407 | 4134 | -0.009105 | 2458 | -0.007995 | ... | 3272.178 | 2923 | 0.009211 | 2209.094 | 1780 | 0.022529 | 1658.026 | 86.558093 | 6.591012 | 100.000000 |
| 1 | ASW_2020_09_28_2 | -0.025427 | 6636 | -0.022822 | 4372 | -0.021184 | 4144 | -0.007454 | 2477 | -0.006254 | ... | 3276.035 | 2942 | 0.007386 | 2218.254 | 1771 | 0.019989 | 1653.686 | 86.043652 | 5.563222 | 99.405669 |
| 2 | ASW_2020_09_28_3 | -0.024675 | 6636 | -0.021948 | 4401 | -0.020278 | 4142 | -0.006614 | 2477 | -0.005435 | ... | 3276.035 | 2942 | 0.007681 | 2218.254 | 1771 | 0.020173 | 1653.686 | 86.876902 | 5.599657 | 100.368318 |
| 3 | ASW_2020_09_28_4 | -0.024353 | 6632 | -0.021656 | 4393 | -0.019961 | 4142 | -0.006287 | 2477 | -0.005064 | ... | 3271.213 | 2923 | 0.007746 | 2209.094 | 1771 | 0.020165 | 1653.686 | 87.117734 | 5.607032 | 100.646549 |
| 4 | ASW_2020_09_28_5 | -0.024163 | 6632 | -0.021448 | 4401 | -0.019691 | 4134 | -0.005975 | 2458 | -0.004768 | ... | 3276.035 | 2942 | 0.007936 | 2218.254 | 1771 | 0.020096 | 1653.686 | 87.298165 | 5.540918 | 100.855000 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 75 | ASW_2020_09_28_76 | 0.033360 | 6633 | 0.035411 | 4154 | 0.035383 | 4135 | 0.040743 | 2479 | 0.041975 | ... | 3226.858 | 2990 | 0.005160 | 2241.396 | 1675 | 0.008315 | 1607.402 | 40.624815 | 2.506664 | 46.933584 |
| 76 | ASW_2020_09_28_77 | 0.039784 | 6633 | 0.040950 | 4154 | 0.040848 | 4077 | 0.043054 | 2479 | 0.043847 | ... | 3225.894 | 3040 | 0.003348 | 2265.502 | 1675 | 0.006200 | 1607.402 | 22.869835 | 1.940948 | 26.421371 |
| 77 | ASW_2020_09_28_78 | 0.043527 | 6335 | 0.043778 | 4163 | 0.043654 | 3962 | 0.043991 | 2345 | 0.043920 | ... | 3224.929 | 2300 | 0.002073 | 1908.730 | 947 | 0.005038 | 1256.416 | 5.198979 | 2.124355 | 6.006347 |
| 78 | ASW_2020_09_28_79 | 0.043639 | 6638 | 0.043704 | 4557 | 0.044042 | 3943 | 0.044228 | 2345 | 0.044163 | ... | 3224.929 | 2321 | 0.002171 | 1918.855 | 947 | 0.005697 | 1256.416 | 3.493992 | 2.447063 | 4.036586 |
| 79 | ASW_2020_09_28_80 | 0.016138 | 6633 | 0.020018 | 4211 | 0.020286 | 4136 | 0.033108 | 2489 | 0.034497 | ... | 3248.072 | 3037 | 0.011578 | 2264.056 | 1698 | 0.018059 | 1618.491 | 102.387413 | 7.200944 | 118.287510 |
80 rows × 31 columns
Purpose of this section is to perform a deep analysis of the sample with respect to the temperature ramp as for example,
plots various scans at different temperature, identify various range of interest.
perform linear regression of desorption data
average scans when isotherm ?
1.2 DATA Cleaning¶
1.2.1 Annealing scan supression and averaging of the isotherms ?¶
1.2.2 Error Analysis¶
After having avaeraged the scan previously, the purpose of this section is to identify the potential sources of errors and to generate error bars for the data accordingly
1.2 DATA Compare¶
1.2.1 MaxA¶
Prepare two dictionnaries¶
Dict 1¶
16-09
i = [0,1,4,7,10,13,16,19,22,25]
j = ["Dep","20","40","60","80","100","120","130","140","160"]
Td = dict(zip(j, i))
nscanTd = len(list(data_anex_df.index.values.tolist()))
print(nscanTd)
print(Td)
31
{'Dep': 0, '20': 1, '40': 4, '60': 7, '80': 10, '100': 13, '120': 16, '130': 19, '140': 22, '160': 25}
Dict 2¶
28-09
i = [0,1,8,16,24,32,40,52,60,79]
j = ["Dep","20","40","60","80","100","120","130","140","160"]
T2d = dict(zip(j, i))
nscanT2d = len(list(data_anex_df_2.index.values.tolist()))
print(nscanT2d)
print(T2d)
80
{'Dep': 0, '20': 1, '40': 8, '60': 16, '80': 24, '100': 32, '120': 40, '130': 52, '140': 60, '160': 79}
Plot¶
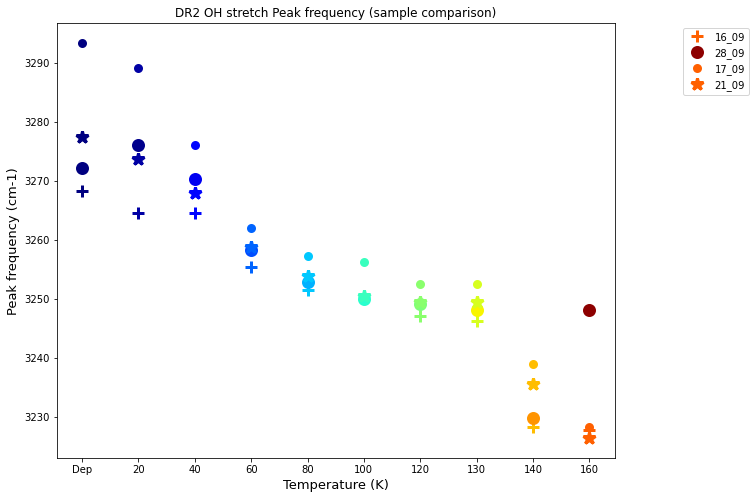
Peak frequency shift¶
fig= plt.figure(figsize=(10,8))
for (k,v), (k2,v2) in zip(Td.items(), T2d.items()):
spl1, = plt.plot(k, data_anex_df.iloc[v,17], '+', mew=3, ms=12, c=cm.jet(v/nscanTd), label= "16_09")
spl2, = plt.plot(k2, data_anex_df_2.iloc[v2,17], 'o', mew=3, ms=10, c=cm.jet(v2/nscanT2d), label= "28_09")
spl3, = plt.plot(k, data_anex_df_3.iloc[v,17], '.', mew=3, ms=12, c=cm.jet(v/nscanTd), label= "17_09")
spl4, = plt.plot(k, data_anex_df_4.iloc[v,17], '*', mew=3, ms=12, c=cm.jet(v/nscanTd), label= "21_09")
plt.title('DR2 OH stretch Peak frequency (sample comparison)')
#plt.axis([3210,3270])
plt.xlabel('Temperature (K)').set_fontsize(13)
plt.ylabel('Peak frequency (cm-1)').set_fontsize(13)
#ax = fig.gca()
#plt.grid()
#plt.legend()
plt.legend([spl1, spl2, spl3, spl4], ["16_09", "28_09", "17_09", "21_09"],bbox_to_anchor =(1.25, 1),)
#plt.savefig('D:\DATA-Processing\PAC\{}/Compare/DR2_{}_{}_PeakA-frequency.png'.format(XP, date, date2))
plt.show()
Integration from normalized data¶
fig= plt.figure(figsize=(10,8))
for (k,v), (k2,v2) in zip(Td.items(), T2d.items()):
spl1, = plt.plot(k, data_anex_df.iloc[v,30], '+', mew=3, ms=12, c=cm.jet(v/nscanTd), label= "16_09")
spl2, = plt.plot(k2, data_anex_df_2.iloc[v2,30], 'o', mew=3, ms=10, c=cm.jet(v2/nscanT2d), label= "28_09")
spl3, = plt.plot(k, data_anex_df_3.iloc[v,30], '.', mew=3, ms=12, c=cm.jet(v/nscanTd), label= "17_09")
spl4, = plt.plot(k, data_anex_df_4.iloc[v,30], '*', mew=3, ms=12, c=cm.jet(v/nscanTd), label= "21_09")
plt.title('DR2 OH stretch Integration value (sample comparison)')
#plt.axis([3210,3270])
plt.xlabel('Temperature (K)').set_fontsize(13)
plt.ylabel('Integration (AU)').set_fontsize(13)
#ax = fig.gca()
#plt.grid()
#plt.legend()
plt.legend([spl1, spl2, spl3, spl4], ["16_09", "28_09", "17_09", "21_09"])
#plt.savefig('D:\DATA-Processing\PAC\{}/Compare/DR2_{}_{}_PeakA-frequency.png'.format(XP, date, date2))
plt.show()
1.3 Data chopping¶
1.3.1 Temperature list splitting¶
Temp = XP_Ramp_df_I.columns[1:].values.tolist()
Temp
['20K ',
'30K',
'40K',
'50K',
'60K',
'70K',
'80K',
'90K',
'100K',
'110K',
'120K',
'125K',
'130K',
'132K',
'134K',
'135K',
'136K',
'137K',
'138K',
'140K',
'142K',
'145K',
'150K',
'155K',
'160K',
'180K',
'200K']
iterate through temp and recover scan number value for T in range 1,2,3,4
T1 = Temp[0:9]
T2 = Temp[8:13]
T3 = Temp[12:20]
T4 = Temp[19:26]
Sanity Check¶
T2
['100K', '110K', '120K', '125K', '130K']
1.1.3 List of sample names per range¶
Range 1¶
z = []
value_1 = []
file_number1 = []
for y in T1:
value = XP_Ramp_df_I.loc[XP_Ramp_df_I.index == date, y].values[0]
if pd.notnull(value):
value_1 = re.findall(r"[-+]?\d*\.\d+|\d+", value)
for items in value_1:
to_plot = str('{}_{}_{}'.format(spl, date, items))
z.append({
'Name' : str(to_plot),
'Temp' : y,
'Date': date,
})
file_number1.append(int(items))
dat1= pd.DataFrame(z)
R1_df = dat1.set_index('Name')
z
[{'Name': 'ASW_2021_05_24_1', 'Temp': '20K ', 'Date': '2021_05_24'},
{'Name': 'ASW_2021_05_24_2', 'Temp': '20K ', 'Date': '2021_05_24'},
{'Name': 'ASW_2021_05_24_3', 'Temp': '20K ', 'Date': '2021_05_24'},
{'Name': 'ASW_2021_05_24_4', 'Temp': '70K', 'Date': '2021_05_24'},
{'Name': 'ASW_2021_05_24_5', 'Temp': '70K', 'Date': '2021_05_24'},
{'Name': 'ASW_2021_05_24_6', 'Temp': '70K', 'Date': '2021_05_24'},
{'Name': 'ASW_2021_05_24_7', 'Temp': '70K', 'Date': '2021_05_24'}]
R1_df
| Temp | Date | |
|---|---|---|
| Name | ||
| ASW_2021_05_24_1 | 20K | 2021_05_24 |
| ASW_2021_05_24_2 | 20K | 2021_05_24 |
| ASW_2021_05_24_3 | 20K | 2021_05_24 |
| ASW_2021_05_24_4 | 70K | 2021_05_24 |
| ASW_2021_05_24_5 | 70K | 2021_05_24 |
| ASW_2021_05_24_6 | 70K | 2021_05_24 |
| ASW_2021_05_24_7 | 70K | 2021_05_24 |
Range 2¶
z = []
value_1 = []
file_number2 = []
for y in T2:
value = XP_Ramp_df_I.loc[XP_Ramp_df_I.index == date, y].values[0]
if pd.notnull(value):
value_1 = re.findall(r"[-+]?\d*\.\d+|\d+", value)
for items in value_1:
to_plot = str('{}_{}_{}'.format(spl, date, items))
z.append({
'Name' : str(to_plot),
'Temp' : y,
'Date': date,
})
file_number2.append(int(items))
dat2= pd.DataFrame(z)
R2_df = dat2.set_index('Name')
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
<ipython-input-21-5bf50306e56a> in <module>
24
25 dat2= pd.DataFrame(z)
---> 26 R2_df = dat2.set_index('Name')
~\Anaconda3\lib\site-packages\pandas\core\frame.py in set_index(self, keys, drop, append, inplace, verify_integrity)
4301
4302 if missing:
-> 4303 raise KeyError(f"None of {missing} are in the columns")
4304
4305 if inplace:
KeyError: "None of ['Name'] are in the columns"
R2_df
Range 3¶
z = []
value_1 = []
file_number3 = []
for y in T3:
value = XP_Ramp_df_I.loc[XP_Ramp_df_I.index == date, y].values[0]
if pd.notnull(value):
value_1 = re.findall(r"[-+]?\d*\.\d+|\d+", value)
for items in value_1:
to_plot = str('{}_{}_{}'.format(spl, date, items))
z.append({
'Name' : str(to_plot),
'Temp' : y,
'Date': date,
})
file_number3.append(int(items))
dat3= pd.DataFrame(z)
R3_df = dat3.set_index('Name')
R3_df
| Temp | Date | |
|---|---|---|
| Name | ||
| ASW_2021_05_24_8 | 135K | 2021_05_24 |
| ASW_2021_05_24_9 | 135K | 2021_05_24 |
| ASW_2021_05_24_10 | 135K | 2021_05_24 |
| ASW_2021_05_24_11 | 135K | 2021_05_24 |
| ASW_2021_05_24_12 | 135K | 2021_05_24 |
| ... | ... | ... |
| ASW_2021_05_24_414 | 140K | 2021_05_24 |
| ASW_2021_05_24_415 | 140K | 2021_05_24 |
| ASW_2021_05_24_416 | 140K | 2021_05_24 |
| ASW_2021_05_24_417 | 140K | 2021_05_24 |
| ASW_2021_05_24_418 | 140K | 2021_05_24 |
411 rows × 2 columns
Range 4¶
z = []
value_1 = []
file_number4 = []
for y in T4:
value = XP_Ramp_df_I.loc[XP_Ramp_df_I.index == date, y].values[0]
if pd.notnull(value):
value_1 = re.findall(r"[-+]?\d*\.\d+|\d+", value)
for items in value_1:
to_plot = str('{}_{}_{}'.format(spl, date, items))
z.append({
'Name' : str(to_plot),
'Temp' : y,
'Date': date,
})
file_number4.append(int(items))
dat4= pd.DataFrame(z)
R4_df = dat4.set_index('Name')
R4_df
| Temp | Date | |
|---|---|---|
| Name | ||
| ASW_2021_05_24_393 | 140K | 2021_05_24 |
| ASW_2021_05_24_394 | 140K | 2021_05_24 |
| ASW_2021_05_24_395 | 140K | 2021_05_24 |
| ASW_2021_05_24_396 | 140K | 2021_05_24 |
| ASW_2021_05_24_397 | 140K | 2021_05_24 |
| ... | ... | ... |
| ASW_2021_05_24_495 | 145K | 2021_05_24 |
| ASW_2021_05_24_496 | 145K | 2021_05_24 |
| ASW_2021_05_24_497 | 145K | 2021_05_24 |
| ASW_2021_05_24_498 | 145K | 2021_05_24 |
| ASW_2021_05_24_499 | 145K | 2021_05_24 |
107 rows × 2 columns
1.4 Plotting¶
Plot 4 subplot in square formating for all 4 T range
1.4.1 T ranges¶
Gridspec¶
gs = gridspec.GridSpec(16, 16,hspace=0.8,wspace=0.5)
fig = plt.figure(figsize=(16,16))
fig.suptitle('20K 10-7 All samples')
ax1 = fig.add_subplot(gs[0:8, 0:8]) # row 0, col 0
nscan = len(list(R1_df['Date'].values.tolist()))
j = 1
for i, j in zip(dat1['Name'], range(1,nscan)):
# for iscan in range(1,nscan):
#DR2
#x = DR2_Allscans_full.Wavenumber
#y = DR2_Allscans_full['{}'.format(i)]
x = DR3_Allscan_df.Wavenumber
y = DR3_Allscan_df['{}'.format(i)]
plt.plot(x,y, label="{}_{}".format(str(R1_df.loc[i]['Date']), str(R1_df.loc[i]['Temp'])), c=cm.jet(j/nscan))
j=+1
ax1.set_title('Full range')
ax1.axis([4000, 2800, 0, 0.5])
plt.legend()
ax2 = fig.add_subplot(gs[0:8, 8:16]) # row 0, col 0
nscan = len(list(R2_df['Date'].values.tolist()))
j = 1
for i, j in zip(dat2['Name'], range(1,nscan)):
# for iscan in range(1,nscan):
#DR2
#x = DR2_Allscans_full.Wavenumber
#y = DR2_Allscans_full['{}'.format(i)]
x = DR3_Allscan_df.Wavenumber
y = DR3_Allscan_df['{}'.format(i)]
plt.plot(x,y, label="{}_{}".format(str(R2_df.loc[i]['Date']), str(R2_df.loc[i]['Temp'])), c=cm.jet(j/nscan))
j=+1
ax2.set_title('Normalized zoom')
ax2.margins(2, 2)
ax2.axis([4000, 2800, 0, 0.5])
plt.legend()
ax3 = fig.add_subplot(gs[8:16, 0:8]) # row 0, col 0
nscan = len(list(R3_df['Date'].values.tolist()))
j = 1
for i, j in zip(dat3['Name'], range(1,nscan)):
# for iscan in range(1,nscan):
#DR2
#x = DR2_Allscans_full.Wavenumber
#y = DR2_Allscans_full['{}'.format(i)]
x = DR3_Allscan_df.Wavenumber
y = DR3_Allscan_df['{}'.format(i)]
plt.plot(x,y, label="{}_{}".format(str(R3_df.loc[i]['Date']), str(R3_df.loc[i]['Temp'])), c=cm.jet(j/nscan))
j=+1
ax3.set_title('BC zoomed')
ax3.axis([4000, 2800, 0, 0.5])
plt.legend()
ax4 = fig.add_subplot(gs[8:16, 8:16]) # row 0, col 0
nscan = len(list(R4_df['Date'].values.tolist()))
j = 1
for i, j in zip(dat4['Name'], range(1,nscan)):
# for iscan in range(1,nscan):
#DR2
#x = DR2_Allscans_full.Wavenumber
#y = DR2_Allscans_full['{}'.format(i)]
x = DR3_Allscan_df.Wavenumber
y = DR3_Allscan_df['{}'.format(i)]
plt.plot(x,y, label="{}_{}".format(str(R4_df.loc[i]['Date']), str(R4_df.loc[i]['Temp'])), c=cm.jet(j/nscan))
j=+1
ax4.set_title('BC zoomed')
ax4.axis([4000, 2800, 0, 0.5])
plt.legend()
plt.show()
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
<ipython-input-26-d01ae63ef9a4> in <module>
30
31 ax2 = fig.add_subplot(gs[0:8, 8:16]) # row 0, col 0
---> 32 nscan = len(list(R2_df['Date'].values.tolist()))
33
34
NameError: name 'R2_df' is not defined
insert condition to supress first scan (ramp) Implement maxA Increase spacing between plots
1.6 Scan Averaging and error analysis ?¶
Supress the first scan and average the remaining scan
Have a though about error bars
1.7 Data processing¶
1.7.1 Linear regression of desorption¶
This is where some fitting options needs to be implemented ie:
linear regression for desorption data
Polynomial for isotherms at 130, 135K ?
1.7.2 polynomial fitting of isotherm at 130K¶
Goal,
\(\color{green}{\text{DP2 : Substraction scan}}\) ¶
use DR2 data
Purpose here is to obtain 4 different substraction spectra for each of the presestablished range (use hot-warm color to plot)
2.1 Data Sanity Check¶
DR2_Allscan_df
| Wavenumber | ASW_2021_05_24_1 | ASW_2021_05_24_2 | ASW_2021_05_24_3 | ASW_2021_05_24_4 | ASW_2021_05_24_5 | ASW_2021_05_24_6 | ASW_2021_05_24_7 | ASW_2021_05_24_8 | ASW_2021_05_24_9 | ... | ASW_2021_05_24_483 | ASW_2021_05_24_484 | ASW_2021_05_24_485 | ASW_2021_05_24_486 | ASW_2021_05_24_487 | ASW_2021_05_24_488 | ASW_2021_05_24_489 | ASW_2021_05_24_490 | ASW_2021_05_24_491 | ASW_2021_05_24_492 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 799.8442 | 6.517882e-02 | 6.611663e-02 | 6.766044e-02 | 7.767758e-02 | 8.016905e-02 | 8.028784e-02 | 0.079780 | 7.776978e-02 | 7.765637e-02 | ... | 0.048800 | 0.048767 | 0.048408 | 0.049178 | 0.047778 | 0.051763 | 0.052555 | 0.055020 | 0.056034 | 0.061171 |
| 1 | 800.3264 | 6.945459e-02 | 6.969134e-02 | 7.088943e-02 | 7.296595e-02 | 7.390441e-02 | 7.500930e-02 | 0.074624 | 7.745363e-02 | 7.897147e-02 | ... | 0.052965 | 0.053671 | 0.052923 | 0.054063 | 0.053984 | 0.055621 | 0.057292 | 0.059009 | 0.059942 | 0.066201 |
| 2 | 800.8085 | 7.292112e-02 | 7.288691e-02 | 7.361745e-02 | 6.737457e-02 | 6.700571e-02 | 6.841946e-02 | 0.068437 | 7.570509e-02 | 7.891863e-02 | ... | 0.056586 | 0.057301 | 0.056603 | 0.057588 | 0.059036 | 0.058832 | 0.060971 | 0.062315 | 0.063480 | 0.070444 |
| 3 | 801.2906 | 6.940653e-02 | 6.975350e-02 | 7.111548e-02 | 6.573625e-02 | 6.443463e-02 | 6.554944e-02 | 0.066081 | 7.037464e-02 | 7.083752e-02 | ... | 0.051400 | 0.051448 | 0.051381 | 0.052487 | 0.053773 | 0.053324 | 0.055295 | 0.055882 | 0.058406 | 0.063919 |
| 4 | 801.7727 | 6.659455e-02 | 6.678519e-02 | 6.828554e-02 | 6.814425e-02 | 6.737859e-02 | 6.831091e-02 | 0.068845 | 6.914572e-02 | 6.787662e-02 | ... | 0.046206 | 0.046325 | 0.046060 | 0.047885 | 0.049168 | 0.048616 | 0.049703 | 0.050537 | 0.053893 | 0.058902 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 6636 | 3998.2570 | 6.052594e-04 | 6.155879e-04 | 5.891753e-04 | 4.509248e-04 | 3.057191e-04 | 3.017703e-04 | 0.000373 | 3.303686e-04 | 5.509026e-04 | ... | 0.000525 | 0.000569 | 0.000513 | 0.000519 | 0.000521 | 0.000546 | 0.000569 | 0.000509 | 0.000523 | 0.000514 |
| 6637 | 3998.7390 | 3.875527e-04 | 3.993715e-04 | 3.591537e-04 | 2.900099e-04 | 1.944961e-04 | 1.994642e-04 | 0.000299 | 2.035522e-04 | 3.159683e-04 | ... | 0.000346 | 0.000352 | 0.000322 | 0.000330 | 0.000345 | 0.000352 | 0.000361 | 0.000334 | 0.000336 | 0.000338 |
| 6638 | 3999.2210 | -8.673617e-19 | -4.336809e-19 | -2.168404e-19 | -3.469447e-18 | -1.301043e-18 | 3.469447e-18 | 0.000121 | 2.168404e-18 | 1.734723e-18 | ... | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| 6639 | 3999.7030 | 7.357823e-04 | 7.429329e-04 | 7.613905e-04 | 3.697233e-04 | 6.489695e-05 | 1.846985e-05 | 0.000126 | 1.990229e-04 | 7.767737e-04 | ... | 0.000430 | 0.000447 | 0.000411 | 0.000422 | 0.000424 | 0.000413 | 0.000430 | 0.000414 | 0.000451 | 0.000435 |
| 6640 | 4000.1850 | 1.665731e-03 | 1.684217e-03 | 1.704210e-03 | 9.846366e-04 | 4.392129e-04 | 3.268367e-04 | 0.000441 | 5.654779e-04 | 1.681778e-03 | ... | 0.001176 | 0.001227 | 0.001154 | 0.001179 | 0.001158 | 0.001152 | 0.001200 | 0.001149 | 0.001240 | 0.001204 |
6641 rows × 493 columns
wavelength = DR2_Allscan_df.T.iloc[0]
wavelength
0 799.8442
1 800.3264
2 800.8085
3 801.2906
4 801.7727
...
6636 3998.2570
6637 3998.7390
6638 3999.2210
6639 3999.7030
6640 4000.1850
Name: Wavenumber, Length: 6641, dtype: float64
2.2 Function definition¶
def substraction(index1, date, index2):
# exp_date1 = input('date of experiment 1 (format YYYY_MM_DD) = ')
# index1 = int(input('index 1 = '))
# exp_date2 = input('date of experiment 2 (format YYYY_MM_DD) = ')
# index2 = int(input('index 2 = '))
data1 = DR2_Allscan_df
#data2 = pd.read_csv('D:\PhD-WS\Projects\PAC\{}}\DATA\{0}\Data\DR2_{}_All_data.csv'.format(XP,date))
data1 = data1.T.iloc[1:].T
#data2 = data2.T.iloc[1:].T
#Scan 1
list_name1 = list(data1.columns.values.tolist())
scan_place1 = 0
for i in range(len(list_name1)):
name_corr = list_name1[i].split('_')
if int(name_corr[-1]) == int(index1):
scan_place1 = i
scan1 = data1.T.iloc[scan_place1]
#Scan 2
list_name2 = list(data1.columns.values.tolist())
scan_place2 = 0
for i in range(len(list_name2)):
name_corr = list_name2[i].split('_')
if int(name_corr[-1]) == int(index2):
scan_place2 = i
scan2 = data1.T.iloc[scan_place2]
sub_scan = numpy.zeros(len(scan1))
for iscan in range(len(sub_scan)):
sub_scan[iscan] = scan2[iscan] - scan1[iscan]
return sub_scan
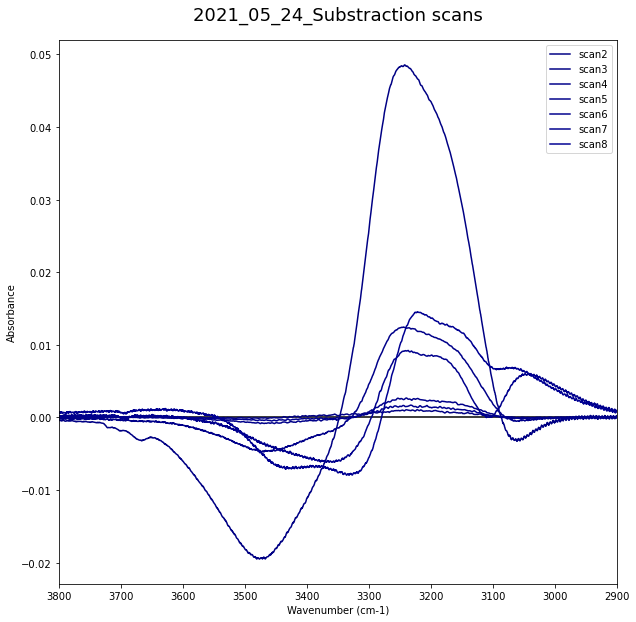
2.3 Plotting¶
All Range¶
#date = "2020_09_15"
data1 = DR2_Allscan_df
data1 = data1.T.iloc[1:].T
#data = data.T.iloc[1:].T
#data = data1
nscan = len(list(data1.columns.values.tolist()))
print(nscan)
#print(data1)
fig = plt.figure(figsize=(16,16))
ax = fig.add_subplot()
ax.set_xlabel('Wavenumber (cm-1)')
ax.set_ylabel('Absorbance')
#ax.set_zlabel('Absorbance')
data_sub = []
data_sub_2 =[]
for iscan in range(nscan-1):
iscan = iscan+1
sub_scan = substraction(iscan, date, iscan+1)
df = pd.DataFrame({'Wavenumber': wavelength, "ASW_"+str(date)+"_" + str(iscan) +"-" + str(iscan+1): sub_scan})
#df.to_csv('D:\PhD-WS\Projects\PAC\XP_1-1\DATA\{}\Data\DP1\DP1_{}_sub-scan_{}.csv'.format(date,date, iscan))
#data_int.append(df)
data_sub.append(
{
'Name' : "ASW_"+str(date)+"_" + str(iscan) +"-" + str(iscan+1),
'Wavenumber' : wavelength,
'sub_scan' : sub_scan,
})
data_sub_2.append(df)
plt.plot(wavelength, sub_scan, c=cm.jet(iscan/nscan), label = "scan{0}".format(str(iscan)))
plt.title('{}_Substraction scans'.format(date), pad=20).set_fontsize(18)
ax.hlines(0,3900,2900)
plt.xlim([3800,2900])
plt.legend()
plt.show()
492
C:\Users\vmrd3\Anaconda3\lib\site-packages\IPython\core\pylabtools.py:132: UserWarning: Creating legend with loc="best" can be slow with large amounts of data.
fig.canvas.print_figure(bytes_io, **kw)

Range 1¶
Insert condition to use only items present in range 1
#date = "2020_09_15"
data1 = DR2_Allscan_df
data1 = data1.T.iloc[1:].T
#data = data.T.iloc[1:].T
#data = data1
nscan = len(list(data1.columns.values.tolist()))
print(nscan)
#print(data1)
fig = plt.figure(figsize=(10,10))
ax = fig.add_subplot()
ax.set_xlabel('Wavenumber (cm-1)')
ax.set_ylabel('Absorbance')
#ax.set_zlabel('Absorbance')
data_sub = []
data_sub_2 =[]
for iscan in file_number1:
iscan = iscan+1
sub_scan = substraction(iscan, date, iscan+1)
df = pd.DataFrame({'Wavenumber': wavelength, "ASW_"+str(date)+"_" + str(iscan) +"-" + str(iscan+1): sub_scan})
#df.to_csv('D:\PhD-WS\Projects\PAC\XP_1-1\DATA\{}\Data\DP1\DP1_{}_sub-scan_{}.csv'.format(date,date, iscan))
#data_int.append(df)
data_sub.append(
{
'Name' : "ASW_"+str(date)+"_" + str(iscan) +"-" + str(iscan+1),
'Wavenumber' : wavelength,
'sub_scan' : sub_scan,
})
data_sub_2.append(df)
plt.plot(wavelength, sub_scan, c=cm.jet(iscan/nscan), label = "scan{0}".format(str(iscan)))
plt.title('{}_Substraction scans'.format(date), pad=20).set_fontsize(18)
ax.hlines(0,3900,2900)
plt.xlim([3800,2900])
plt.legend()
plt.show()
492
df
| Wavenumber | ASW_2021_05_24_8-9 | |
|---|---|---|
| 0 | 799.8442 | -1.134107e-04 |
| 1 | 800.3264 | 1.517846e-03 |
| 2 | 800.8085 | 3.213542e-03 |
| 3 | 801.2906 | 4.628880e-04 |
| 4 | 801.7727 | -1.269106e-03 |
| ... | ... | ... |
| 6636 | 3998.2570 | 2.205339e-04 |
| 6637 | 3998.7390 | 1.124161e-04 |
| 6638 | 3999.2210 | -4.336809e-19 |
| 6639 | 3999.7030 | 5.777508e-04 |
| 6640 | 4000.1850 | 1.116301e-03 |
6641 rows × 2 columns
data_sub
[{'Name': 'ASW_2021_05_24_2-3',
'Wavenumber': 0 799.8442
1 800.3264
2 800.8085
3 801.2906
4 801.7727
...
6636 3998.2570
6637 3998.7390
6638 3999.2210
6639 3999.7030
6640 4000.1850
Name: Wavenumber, Length: 6641, dtype: float64,
'sub_scan': array([1.54380954e-03, 1.19808933e-03, 7.30539117e-04, ...,
2.16840434e-19, 1.84575997e-05, 1.99932794e-05])},
{'Name': 'ASW_2021_05_24_3-4',
'Wavenumber': 0 799.8442
1 800.3264
2 800.8085
3 801.2906
4 801.7727
...
6636 3998.2570
6637 3998.7390
6638 3999.2210
6639 3999.7030
6640 4000.1850
Name: Wavenumber, Length: 6641, dtype: float64,
'sub_scan': array([ 1.00171360e-02, 2.07651706e-03, -6.24288189e-03, ...,
-3.25260652e-18, -3.91667293e-04, -7.19573885e-04])},
{'Name': 'ASW_2021_05_24_4-5',
'Wavenumber': 0 799.8442
1 800.3264
2 800.8085
3 801.2906
4 801.7727
...
6636 3998.2570
6637 3998.7390
6638 3999.2210
6639 3999.7030
6640 4000.1850
Name: Wavenumber, Length: 6641, dtype: float64,
'sub_scan': array([ 2.49147249e-03, 9.38463666e-04, -3.68855160e-04, ...,
2.16840434e-18, -3.04826309e-04, -5.45423718e-04])},
{'Name': 'ASW_2021_05_24_5-6',
'Wavenumber': 0 799.8442
1 800.3264
2 800.8085
3 801.2906
4 801.7727
...
6636 3998.2570
6637 3998.7390
6638 3999.2210
6639 3999.7030
6640 4000.1850
Name: Wavenumber, Length: 6641, dtype: float64,
'sub_scan': array([ 1.18793290e-04, 1.10489031e-03, 1.41374733e-03, ...,
4.77048956e-18, -4.64270998e-05, -1.12376200e-04])},
{'Name': 'ASW_2021_05_24_6-7',
'Wavenumber': 0 799.8442
1 800.3264
2 800.8085
3 801.2906
4 801.7727
...
6636 3998.2570
6637 3998.7390
6638 3999.2210
6639 3999.7030
6640 4000.1850
Name: Wavenumber, Length: 6641, dtype: float64,
'sub_scan': array([-5.07472153e-04, -3.85217634e-04, 1.70768859e-05, ...,
1.20905107e-04, 1.07518578e-04, 1.13815049e-04])},
{'Name': 'ASW_2021_05_24_7-8',
'Wavenumber': 0 799.8442
1 800.3264
2 800.8085
3 801.2906
4 801.7727
...
6636 3998.2570
6637 3998.7390
6638 3999.2210
6639 3999.7030
6640 4000.1850
Name: Wavenumber, Length: 6641, dtype: float64,
'sub_scan': array([-2.01059091e-03, 2.82954489e-03, 7.26855070e-03, ...,
-1.20905107e-04, 7.30345003e-05, 1.24826107e-04])},
{'Name': 'ASW_2021_05_24_8-9',
'Wavenumber': 0 799.8442
1 800.3264
2 800.8085
3 801.2906
4 801.7727
...
6636 3998.2570
6637 3998.7390
6638 3999.2210
6639 3999.7030
6640 4000.1850
Name: Wavenumber, Length: 6641, dtype: float64,
'sub_scan': array([-1.13410710e-04, 1.51784552e-03, 3.21354176e-03, ...,
-4.33680869e-19, 5.77750781e-04, 1.11630056e-03])}]
data_sub_2
[ Wavenumber ASW_2021_05_24_2-3
0 799.8442 1.543810e-03
1 800.3264 1.198089e-03
2 800.8085 7.305391e-04
3 801.2906 1.361979e-03
4 801.7727 1.500349e-03
... ... ...
6636 3998.2570 -2.641260e-05
6637 3998.7390 -4.021780e-05
6638 3999.2210 2.168404e-19
6639 3999.7030 1.845760e-05
6640 4000.1850 1.999328e-05
[6641 rows x 2 columns],
Wavenumber ASW_2021_05_24_3-4
0 799.8442 1.001714e-02
1 800.3264 2.076517e-03
2 800.8085 -6.242882e-03
3 801.2906 -5.379231e-03
4 801.7727 -1.412898e-04
... ... ...
6636 3998.2570 -1.382505e-04
6637 3998.7390 -6.914371e-05
6638 3999.2210 -3.252607e-18
6639 3999.7030 -3.916673e-04
6640 4000.1850 -7.195739e-04
[6641 rows x 2 columns],
Wavenumber ASW_2021_05_24_4-5
0 799.8442 2.491472e-03
1 800.3264 9.384637e-04
2 800.8085 -3.688552e-04
3 801.2906 -1.301614e-03
4 801.7727 -7.656528e-04
... ... ...
6636 3998.2570 -1.452057e-04
6637 3998.7390 -9.551389e-05
6638 3999.2210 2.168404e-18
6639 3999.7030 -3.048263e-04
6640 4000.1850 -5.454237e-04
[6641 rows x 2 columns],
Wavenumber ASW_2021_05_24_5-6
0 799.8442 1.187933e-04
1 800.3264 1.104890e-03
2 800.8085 1.413747e-03
3 801.2906 1.114804e-03
4 801.7727 9.323114e-04
... ... ...
6636 3998.2570 -3.948800e-06
6637 3998.7390 4.968100e-06
6638 3999.2210 4.770490e-18
6639 3999.7030 -4.642710e-05
6640 4000.1850 -1.123762e-04
[6641 rows x 2 columns],
Wavenumber ASW_2021_05_24_6-7
0 799.8442 -0.000507
1 800.3264 -0.000385
2 800.8085 0.000017
3 801.2906 0.000531
4 801.7727 0.000534
... ... ...
6636 3998.2570 0.000071
6637 3998.7390 0.000099
6638 3999.2210 0.000121
6639 3999.7030 0.000108
6640 4000.1850 0.000114
[6641 rows x 2 columns],
Wavenumber ASW_2021_05_24_7-8
0 799.8442 -0.002011
1 800.3264 0.002830
2 800.8085 0.007269
3 801.2906 0.004294
4 801.7727 0.000301
... ... ...
6636 3998.2570 -0.000042
6637 3998.7390 -0.000095
6638 3999.2210 -0.000121
6639 3999.7030 0.000073
6640 4000.1850 0.000125
[6641 rows x 2 columns],
Wavenumber ASW_2021_05_24_8-9
0 799.8442 -1.134107e-04
1 800.3264 1.517846e-03
2 800.8085 3.213542e-03
3 801.2906 4.628880e-04
4 801.7727 -1.269106e-03
... ... ...
6636 3998.2570 2.205339e-04
6637 3998.7390 1.124161e-04
6638 3999.2210 -4.336809e-19
6639 3999.7030 5.777508e-04
6640 4000.1850 1.116301e-03
[6641 rows x 2 columns]]
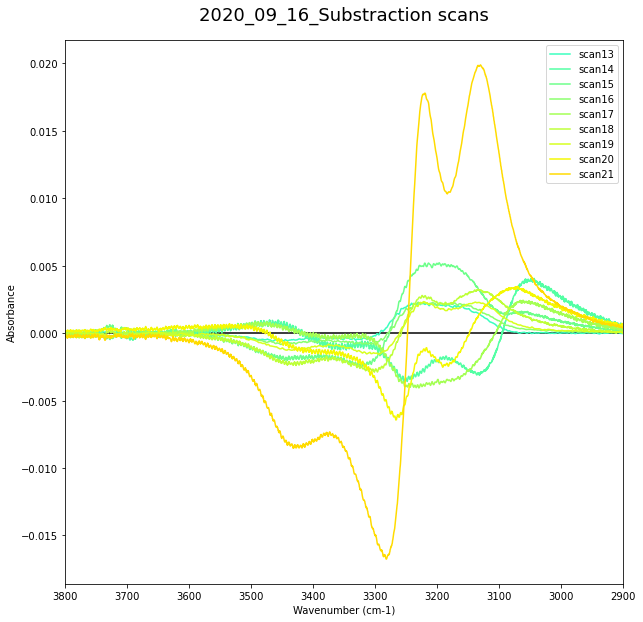
Range 2¶
#date = "2020_09_15"
data1 = DR2_Allscan_df
data1 = data1.T.iloc[1:].T
#data = data.T.iloc[1:].T
#data = data1
nscan = len(list(data1.columns.values.tolist()))
print(nscan)
#print(data1)
fig = plt.figure(figsize=(10,10))
ax = fig.add_subplot()
ax.set_xlabel('Wavenumber (cm-1)')
ax.set_ylabel('Absorbance')
#ax.set_zlabel('Absorbance')
data_sub = []
data_sub_2 =[]
for iscan in file_number2:
iscan = iscan+1
sub_scan = substraction(iscan, date, iscan+1)
df = pd.DataFrame({'Wavenumber': wavelength, "ASW_"+str(date)+"_" + str(iscan) +"-" + str(iscan+1): sub_scan})
#df.to_csv('D:\PhD-WS\Projects\PAC\XP_1-1\DATA\{}\Data\DP1\DP1_{}_sub-scan_{}.csv'.format(date,date, iscan))
#data_int.append(df)
data_sub.append(
{
'Name' : "ASW_"+str(date)+"_" + str(iscan) +"-" + str(iscan+1),
'Wavenumber' : wavelength,
'sub_scan' : sub_scan,
})
data_sub_2.append(df)
plt.plot(wavelength, sub_scan, c=cm.jet(iscan/nscan), label = "scan{0}".format(str(iscan)))
plt.title('{}_Substraction scans'.format(date), pad=20).set_fontsize(18)
ax.hlines(0,3900,2900)
plt.xlim([3800,2900])
plt.legend()
plt.show()
31
Range 3¶
#date = "2020_09_15"
data1 = DR2_Allscan_df
data1 = data1.T.iloc[1:].T
#data = data.T.iloc[1:].T
#data = data1
nscan = len(list(data1.columns.values.tolist()))
print(nscan)
#print(data1)
fig = plt.figure(figsize=(10,10))
ax = fig.add_subplot()
ax.set_xlabel('Wavenumber (cm-1)')
ax.set_ylabel('Absorbance')
#ax.set_zlabel('Absorbance')
data_sub = []
data_sub_2 =[]
for iscan in file_number3:
iscan = iscan+1
sub_scan = substraction(iscan, date, iscan+1)
df = pd.DataFrame({'Wavenumber': wavelength, "ASW_"+str(date)+"_" + str(iscan) +"-" + str(iscan+1): sub_scan})
#df.to_csv('D:\PhD-WS\Projects\PAC\XP_1-1\DATA\{}\Data\DP1\DP1_{}_sub-scan_{}.csv'.format(date,date, iscan))
#data_int.append(df)
data_sub.append(
{
'Name' : "ASW_"+str(date)+"_" + str(iscan) +"-" + str(iscan+1),
'Wavenumber' : wavelength,
'sub_scan' : sub_scan,
})
data_sub_2.append(df)
plt.plot(wavelength, sub_scan, c=cm.jet(iscan/nscan), label = "scan{0}".format(str(iscan)))
plt.title('{}_Substraction scans'.format(date), pad=20).set_fontsize(18)
ax.hlines(0,3900,2900)
plt.xlim([3800,2900])
plt.ylim([-0.0075,0.0075])
#plt.legend()
plt.show()
492
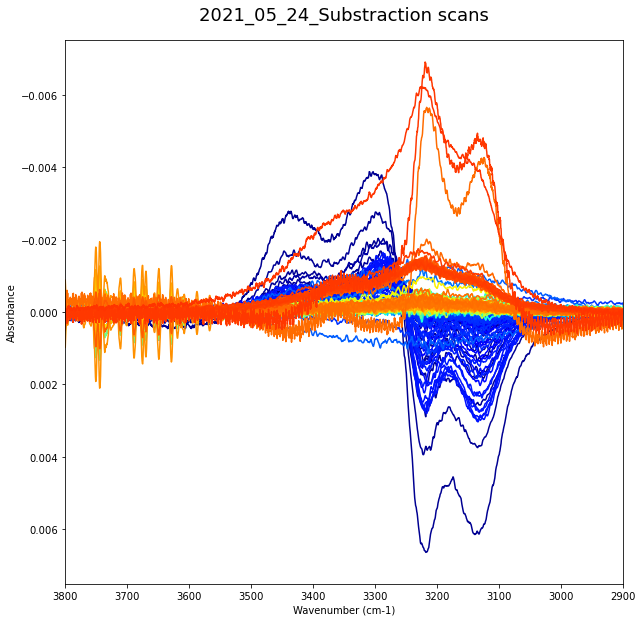
Range 4¶
#date = "2020_09_15"
data1 = DR2_Allscan_df
data1 = data1.T.iloc[1:].T
#data = data.T.iloc[1:].T
#data = data1
nscan = len(list(data1.columns.values.tolist()))
print(nscan)
#print(data1)
fig = plt.figure(figsize=(10,10))
ax = fig.add_subplot()
ax.set_xlabel('Wavenumber (cm-1)')
ax.set_ylabel('Absorbance')
#ax.set_zlabel('Absorbance')
data_sub = []
data_sub_2 =[]
for iscan in file_number4:
iscan = iscan+1
sub_scan = substraction(iscan, date, iscan+1)
df = pd.DataFrame({'Wavenumber': wavelength, "ASW_"+str(date)+"_" + str(iscan) +"-" + str(iscan+1): sub_scan})
#df.to_csv('D:\PhD-WS\Projects\PAC\XP_1-1\DATA\{}\Data\DP1\DP1_{}_sub-scan_{}.csv'.format(date,date, iscan))
#data_int.append(df)
data_sub.append(
{
'Name' : "ASW_"+str(date)+"_" + str(iscan) +"-" + str(iscan+1),
'Wavenumber' : wavelength,
'sub_scan' : sub_scan,
})
data_sub_2.append(df)
plt.plot(wavelength, sub_scan, c=cm.jet(iscan/nscan), label = "scan{0}".format(str(iscan)))
plt.title('{}_Substraction scans'.format(date), pad=20).set_fontsize(18)
ax.hlines(0,3900,2900)
plt.xlim([3800,2900])
plt.legend()
plt.show()
492
Preliminary analysis¶
Purpose here would be (for the different physical processes that the ice undergo upon annealing) to investigate the different contributions lost and gain from the substraction spectra
This will subsequently be used as initial guesses for the gaussian fit of the whole samples
An intermediate gaussian fitting routine could be implemented here with the substraction spectra
Export Data Sub as csv¶
Concatenation¶
data_sub
data_sub_2_df = reduce(lambda left,right: pd.merge(left,right, on=['Wavenumber'],how='outer'), data_sub_2)
Export as csv¶
\(\color{green}{\text{DP3 : Gaussian Fitting}}\) ¶
Gaussian fit 1¶
x = DR2_A_df["Wavenumber"]
y = DR2_A_df["ASW_2020_09_16_1"]
pars = exp_mod.guess(y, x=x)
gauss1 = GaussianModel(prefix='g1_')
pars.update(gauss1.make_params())
pars['g1_center'].set(value=3400, min=3300, max=3500)
pars['g1_sigma'].set(value=15, min=3)
pars['g1_amplitude'].set(value=10, min=1)
gauss2 = GaussianModel(prefix='g2_')
pars.update(gauss2.make_params())
pars['g2_center'].set(value=3260, min=3200, max=3300)
pars['g2_sigma'].set(value=15, min=3)
pars['g2_amplitude'].set(value=10, min=1)
mod = gauss1 + gauss2
init = mod.eval(pars, x=x)
out = mod.fit(y, pars, x=x)
print(out.fit_report(min_correl=0.5))
fig, axes = plt.subplots(1, 2, figsize=(12.8, 4.8))
axes[0].plot(x, y, 'b')
axes[0].plot(x, init, 'k--', label='initial fit')
axes[0].plot(x, out.best_fit, 'r-', label='best fit')
axes[0].invert_xaxis()
axes[0].legend(loc='best')
comps = out.eval_components(x=x)
axes[1].plot(x, y, 'b')
axes[1].plot(x, comps['g1_'], 'g--', label='Gaussian component 1')
axes[1].plot(x, comps['g2_'], 'm--', label='Gaussian component 2')
axes[1].invert_xaxis()
axes[1].legend(loc='best')
plt.show()
# <end examples/doc_builtinmodels_nistgauss.py>
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
<ipython-input-42-82c111bdc377> in <module>
----> 1 x = DR2_A_df["Wavenumber"]
2 y = DR2_A_df["ASW_2020_09_16_1"]
3
4 pars = exp_mod.guess(y, x=x)
5
NameError: name 'DR2_A_df' is not defined